SEM (notes for myself)
p.21
Covariance Derivation
The covariance between two random variables and is defined as:
Step 1: Expanding the expression
First, expand the product inside the expectation:
Step 2: Taking the expectation
Now, take the expectation of both sides:
Step 3: Simplifying the result
Notice that the terms cancel out:
Thus, we arrive at the final formula for the covariance:
p.22
Covariance Derivation in Confirmatory Factor Analysis
We start with the observed variables and :
We want to find :
Using bilinearity of covariance:
Assuming:
Then:
p.23
1. Derivation: Cov(x, c′) = 0
Let be a random vector and a constant vector.
But since is constant, , so:
Hence:
2. Derivation: Var(x) = Cov(x, x′) = Σ
By definition:
This is the population covariance matrix, denoted as:
3. Numerical Example in R
# Set seed and generate data
set.seed(123)
# Simulate random vector x (5 observations, 3 variables)
x <- matrix(c(1, 2, 3, 4, 5,
2, 3, 4, 5, 6,
5, 4, 3, 2, 1), ncol = 3)
x
## [,1] [,2] [,3]
## [1,] 1 2 5
## [2,] 2 3 4
## [3,] 3 4 3
## [4,] 4 5 2
## [5,] 5 6 1
x_centered <- scale(x, center = TRUE, scale = FALSE)
x_centered
## [,1] [,2] [,3]
## [1,] -2 -2 2
## [2,] -1 -1 1
## [3,] 0 0 0
## [4,] 1 1 -1
## [5,] 2 2 -2
## attr(,"scaled:center")
## [1] 3 4 3
4. Confirm Var(x) = Cov(x, x′)
# Compute covariance matrix of x manually
# population covariance matrix
Sigma_manual_p <- t(x_centered) %*% x_centered / (nrow(x))
Sigma_manual_p
## [,1] [,2] [,3]
## [1,] 2 2 -2
## [2,] 2 2 -2
## [3,] -2 -2 2
# sample covariance matrix
Sigma_manual_s <- t(x_centered) %*% x_centered / (nrow(x) - 1)
Sigma_manual_s
## [,1] [,2] [,3]
## [1,] 2.5 2.5 -2.5
## [2,] 2.5 2.5 -2.5
## [3,] -2.5 -2.5 2.5
# Compare with built-in cov()
# sample covariance matrix
Sigma_builtin <- cov(x)
Sigma_builtin
## [,1] [,2] [,3]
## [1,] 2.5 2.5 -2.5
## [2,] 2.5 2.5 -2.5
## [3,] -2.5 -2.5 2.5
p.28
Step 1: Create the data for Disposable Income and Consumers’ Expenditures
income <- c(433, 483, 479, 486, 494, 498, 511, 534, 478, 440, 372, 381, 419, 449, 511, 520, 477, 517, 548, 629)
consum <- c(394, 423, 437, 434, 447, 447, 466, 474, 439, 399, 350, 364, 392, 416, 463, 469, 444, 471, 494, 529)
Step 2: Combine the data into a matrix
data <- cbind(consum, income)
data
## consum income
## [1,] 394 433
## [2,] 423 483
## [3,] 437 479
## [4,] 434 486
## [5,] 447 494
## [6,] 447 498
## [7,] 466 511
## [8,] 474 534
## [9,] 439 478
## [10,] 399 440
## [11,] 350 372
## [12,] 364 381
## [13,] 392 419
## [14,] 416 449
## [15,] 463 511
## [16,] 469 520
## [17,] 444 477
## [18,] 471 517
## [19,] 494 548
## [20,] 529 629
Step 3: Center the data by subtracting the mean of each variable
Z <- scale(data, center = TRUE, scale = FALSE) # Centers the data (but doesn't scale it)
Z
## consum income
## [1,] -43.6 -49.95
## [2,] -14.6 0.05
## [3,] -0.6 -3.95
## [4,] -3.6 3.05
## [5,] 9.4 11.05
## [6,] 9.4 15.05
## [7,] 28.4 28.05
## [8,] 36.4 51.05
## [9,] 1.4 -4.95
## [10,] -38.6 -42.95
## [11,] -87.6 -110.95
## [12,] -73.6 -101.95
## [13,] -45.6 -63.95
## [14,] -21.6 -33.95
## [15,] 25.4 28.05
## [16,] 31.4 37.05
## [17,] 6.4 -5.95
## [18,] 33.4 34.05
## [19,] 56.4 65.05
## [20,] 91.4 146.05
## attr(,"scaled:center")
## consum income
## 437.60 482.95
Step 4: Compute Z’ (transpose of Z)
Z_t <- t(Z)
Z_t
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11]
## consum -43.60 -14.60 -0.60 -3.60 9.40 9.40 28.40 36.40 1.40 -38.60 -87.60
## income -49.95 0.05 -3.95 3.05 11.05 15.05 28.05 51.05 -4.95 -42.95 -110.95
## [,12] [,13] [,14] [,15] [,16] [,17] [,18] [,19] [,20]
## consum -73.60 -45.60 -21.60 25.40 31.40 6.40 33.40 56.40 91.40
## income -101.95 -63.95 -33.95 28.05 37.05 -5.95 34.05 65.05 146.05
## attr(,"scaled:center")
## consum income
## 437.60 482.95
Step 5: Compute Z’Z (matrix multiplication of Z’ and Z)
Z_t_Z <- Z_t %*% Z
Z_t_Z
## consum income
## consum 35886.8 47584.60
## income 47584.6 64992.95
Step 6: Apply the 1/(N-1) factor
N <- nrow(Z) # Number of observations (rows)
cov_matrix_manual <- (1 / (N - 1)) * Z_t_Z
# Display the result
print(cov_matrix_manual)
## consum income
## consum 1888.779 2504.453
## income 2504.453 3420.682
p.28
Introduction
In multivariate statistics, it is often important to detect observations that deviate significantly from the center of the multivariate data cloud. One useful tool is the matrix:
Where:
is the mean-centered data matrix of size ,
is the cross-product matrix,
is its inverse,
is a square matrix whose diagonal entries represent the multivariate “distance” of each observation from the center.
The average value of is:
Observations with much higher values than the average are potential multivariate outliers.
Step-by-Step Example
We will use a simple example dataset with 3 observations and 2 variables.
# Define the data matrix (3 observations, 2 variables)
X <- matrix(c(2, 3, 4, 4, 6, 5), ncol = 2, byrow = FALSE)
colnames(X) <- c("X1", "X2")
rownames(X) <- paste0("Obs", 1:3)
X
## X1 X2
## Obs1 2 4
## Obs2 3 6
## Obs3 4 5
Step 1: Center the Data (Create Z)
We subtract the mean from each variable to obtain the matrix .
Z <- scale(X, center = TRUE, scale = FALSE)
Z
## X1 X2
## Obs1 -1 -1
## Obs2 0 1
## Obs3 1 0
## attr(,"scaled:center")
## X1 X2
## 3 5
Step 2: Compute
ZtZ <- t(Z) %*% Z
ZtZ
## X1 X2
## X1 2 1
## X2 1 2
Step 3: Compute
ZtZ_inv <- solve(ZtZ)
ZtZ_inv
## X1 X2
## X1 0.6666667 -0.3333333
## X2 -0.3333333 0.6666667
Step 4: Compute
A <- Z %*% ZtZ_inv %*% t(Z)
round(A, 3)
## Obs1 Obs2 Obs3
## Obs1 0.667 -0.333 -0.333
## Obs2 -0.333 0.667 -0.333
## Obs3 -0.333 -0.333 0.667
Step 5: Extract Diagonal Elements
These diagonal values represent the multivariate distance for each observation.
a_ii <- diag(A)
names(a_ii) <- rownames(X)
a_ii
## Obs1 Obs2 Obs3
## 0.6666667 0.6666667 0.6666667
Step 6: Compare to Expected Average
p_plus_q <- ncol(Z) # total number of observed variables (p + q)
N <- nrow(Z) # number of observations
expected_mean <- p_plus_q / N
expected_mean
## [1] 0.6666667
Any significantly greater than 0.6666667 may indicate a multivariate outlier.
Step 7: Visualization
barplot(a_ii, names.arg = names(a_ii),
main = "Multivariate Distance (a_ii) for Each Observation",
ylab = "a_ii", col = "skyblue", ylim = c(0, 1))
abline(h = expected_mean, col = "red", lty = 2)
legend("topright", legend = paste("Expected Mean =", round(expected_mean, 3)),
col = "red", lty = 2)
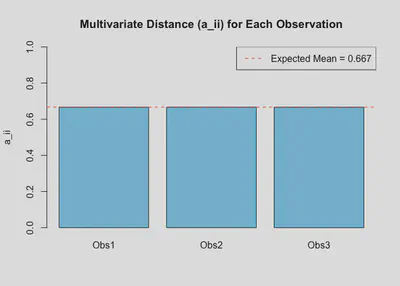
Conclusion
- Matrix provides a way to measure the multivariate distance of each observation.
- Diagonal values indicate how far each case is from the multivariate mean.
- The average of the ’s is , which provides a benchmark.
- Observations with high values are flagged as potential outliers in multivariate space.
This method is especially helpful in the context of SEM, factor analysis, or other multivariate procedures where unusual cases may affect model fit or estimates.
p. 30
Introduction
The formula for calculating multidimensional outliers:
Where:
is the standardized data matrix.
is the product of the transposed matrix and itself.
is the inverse of the matrix .
The final result gives a measure of how far each observation is from the multivariate centroid.
We will apply this formula to a dataset that contains estimates of cloud cover (COVER1, COVER2, COVER3).
Data Preparation
# Define the data
data <- data.frame(
COVER1 = c(0, 20, 80, 50, 5, 1, 5, 0, 10, 0, 0, 10, 0, 10, 0, 0, 5, 10, 20, 35, 90, 50, 35, 25, 0, 0, 10, 40, 35, 55, 35, 0, 0, 5, 20, 0, 0, 0, 15, 95, 40, 40, 15, 30, 75, 100, 100, 100, 100, 100, 100, 100, 0, 5, 80, 80, 80, 40, 20, 1),
COVER2 = c(5, 20, 85, 50, 2, 1, 5, 0, 15, 0, 0, 30, 2, 10, 0, 0, 0, 20, 45, 75, 99, 90, 85, 15, 0, 0, 10, 75, 70, 90, 95, 0, 0, 1, 60, 0, 0, 0, 55, 0, 35, 50, 60, 30, 85, 100, 90, 95, 95, 99, 30, 5, 0, 5, 90, 95, 90, 55, 40, 0),
COVER3 = c(0, 20, 90, 70, 5, 2, 2, 0, 5, 0, 0, 10, 2, 5, 0, 0, 20, 20, 15, 60, 100, 80, 70, 40, 0, 0, 20, 30, 20, 90, 80, 0, 0, 2, 50, 0, 0, 0, 50, 40, 30, 40, 5, 15, 75, 100, 85, 100, 100, 100, 95, 95, 0, 5, 85, 80, 70, 50, 5, 0)
)
dplyr::glimpse(data)
## Rows: 60
## Columns: 3
## $ COVER1 <dbl> 0, 20, 80, 50, 5, 1, 5, 0, 10, 0, 0, 10, 0, 10, 0, 0, 5, 10, 20…
## $ COVER2 <dbl> 5, 20, 85, 50, 2, 1, 5, 0, 15, 0, 0, 30, 2, 10, 0, 0, 0, 20, 45…
## $ COVER3 <dbl> 0, 20, 90, 70, 5, 2, 2, 0, 5, 0, 0, 10, 2, 5, 0, 0, 20, 20, 15,…
# View the data to check it
head(data)
## COVER1 COVER2 COVER3
## 1 0 5 0
## 2 20 20 20
## 3 80 85 90
## 4 50 50 70
## 5 5 2 5
## 6 1 1 2
Mean-Centering
We will now mean-center the data to create the Z matrix.
# Mean-centering the data (subtracting the mean of each column from the data)
Z <- scale(data, center = TRUE, scale = FALSE)
Z
## COVER1 COVER2 COVER3
## [1,] -32.95 -32.65 -35.55
## [2,] -12.95 -17.65 -15.55
## [3,] 47.05 47.35 54.45
## [4,] 17.05 12.35 34.45
## [5,] -27.95 -35.65 -30.55
## [6,] -31.95 -36.65 -33.55
## [7,] -27.95 -32.65 -33.55
## [8,] -32.95 -37.65 -35.55
## [9,] -22.95 -22.65 -30.55
## [10,] -32.95 -37.65 -35.55
## [11,] -32.95 -37.65 -35.55
## [12,] -22.95 -7.65 -25.55
## [13,] -32.95 -35.65 -33.55
## [14,] -22.95 -27.65 -30.55
## [15,] -32.95 -37.65 -35.55
## [16,] -32.95 -37.65 -35.55
## [17,] -27.95 -37.65 -15.55
## [18,] -22.95 -17.65 -15.55
## [19,] -12.95 7.35 -20.55
## [20,] 2.05 37.35 24.45
## [21,] 57.05 61.35 64.45
## [22,] 17.05 52.35 44.45
## [23,] 2.05 47.35 34.45
## [24,] -7.95 -22.65 4.45
## [25,] -32.95 -37.65 -35.55
## [26,] -32.95 -37.65 -35.55
## [27,] -22.95 -27.65 -15.55
## [28,] 7.05 37.35 -5.55
## [29,] 2.05 32.35 -15.55
## [30,] 22.05 52.35 54.45
## [31,] 2.05 57.35 44.45
## [32,] -32.95 -37.65 -35.55
## [33,] -32.95 -37.65 -35.55
## [34,] -27.95 -36.65 -33.55
## [35,] -12.95 22.35 14.45
## [36,] -32.95 -37.65 -35.55
## [37,] -32.95 -37.65 -35.55
## [38,] -32.95 -37.65 -35.55
## [39,] -17.95 17.35 14.45
## [40,] 62.05 -37.65 4.45
## [41,] 7.05 -2.65 -5.55
## [42,] 7.05 12.35 4.45
## [43,] -17.95 22.35 -30.55
## [44,] -2.95 -7.65 -20.55
## [45,] 42.05 47.35 39.45
## [46,] 67.05 62.35 64.45
## [47,] 67.05 52.35 49.45
## [48,] 67.05 57.35 64.45
## [49,] 67.05 57.35 64.45
## [50,] 67.05 61.35 64.45
## [51,] 67.05 -7.65 59.45
## [52,] 67.05 -32.65 59.45
## [53,] -32.95 -37.65 -35.55
## [54,] -27.95 -32.65 -30.55
## [55,] 47.05 52.35 49.45
## [56,] 47.05 57.35 44.45
## [57,] 47.05 52.35 34.45
## [58,] 7.05 17.35 14.45
## [59,] -12.95 2.35 -30.55
## [60,] -31.95 -37.65 -35.55
## attr(,"scaled:center")
## COVER1 COVER2 COVER3
## 32.95 37.65 35.55
# Check the mean-centered data (Z matrix)
head(Z)
## COVER1 COVER2 COVER3
## [1,] -32.95 -32.65 -35.55
## [2,] -12.95 -17.65 -15.55
## [3,] 47.05 47.35 54.45
## [4,] 17.05 12.35 34.45
## [5,] -27.95 -35.65 -30.55
## [6,] -31.95 -36.65 -33.55
Calculating
Next, we compute the product of the transposed matrix and the matrix .
# Calculate Z'Z (transposed Z matrix multiplied by Z)
ZZT <- t(Z) %*% Z
# View the result of Z'Z
ZZT
## COVER1 COVER2 COVER3
## COVER1 76734.85 60191.95 72989.65
## COVER2 60191.95 86335.65 70795.55
## COVER3 72989.65 70795.55 82812.85
Inverse of
Now, we calculate the inverse of the matrix .
# Calculate the inverse of Z'Z
ZZT_inv <- solve(ZZT)
# View the inverse of Z'Z
ZZT_inv
## COVER1 COVER2 COVER3
## COVER1 8.186873e-05 6.996057e-06 -7.813835e-05
## COVER2 6.996057e-06 3.933723e-05 -3.979504e-05
## COVER3 -7.813835e-05 -3.979504e-05 1.149653e-04
Calculate
Finally, using the inverse of , we compute the multidimensional outlier scores.
# Calculate the multidimensional outliers
outliers <- Z %*% ZZT_inv %*% t(Z)
# Extract the diagonal of the outlier matrix (outlier scores)
outlier_scores <- diag(outliers)
# View the outlier scores for each observation
outlier_scores
## [1] 0.015726795 0.003667134 0.035888471 0.043531791 0.015067469 0.016818904
## [7] 0.014336636 0.017711895 0.013230015 0.017711895 0.017711895 0.015735922
## [13] 0.016767037 0.012571539 0.017711895 0.017711895 0.047722524 0.011226588
## [19] 0.033505745 0.044503013 0.051723149 0.067601060 0.085473686 0.043702080
## [25] 0.017711895 0.017711895 0.019881286 0.088784132 0.115257068 0.090112579
## [31] 0.141386847 0.017711895 0.017711895 0.016124246 0.056874143 0.017711895
## [37] 0.017711895 0.017711895 0.078447969 0.310744264 0.012569282 0.004286889
## [43] 0.116357798 0.029894426 0.031821437 0.061860674 0.081910183 0.059274389
## [49] 0.059274389 0.061186068 0.182763526 0.317232262 0.017711895 0.013128462
## [55] 0.034993660 0.045793479 0.063100245 0.005752862 0.064705791 0.017427449
sum(outlier_scores)
## [1] 3
Sample Covariance Matrix
Z_t <- t(Z)
Z_t
## [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
## COVER1 -32.95 -12.95 47.05 17.05 -27.95 -31.95 -27.95 -32.95 -22.95 -32.95
## COVER2 -32.65 -17.65 47.35 12.35 -35.65 -36.65 -32.65 -37.65 -22.65 -37.65
## COVER3 -35.55 -15.55 54.45 34.45 -30.55 -33.55 -33.55 -35.55 -30.55 -35.55
## [,11] [,12] [,13] [,14] [,15] [,16] [,17] [,18] [,19] [,20]
## COVER1 -32.95 -22.95 -32.95 -22.95 -32.95 -32.95 -27.95 -22.95 -12.95 2.05
## COVER2 -37.65 -7.65 -35.65 -27.65 -37.65 -37.65 -37.65 -17.65 7.35 37.35
## COVER3 -35.55 -25.55 -33.55 -30.55 -35.55 -35.55 -15.55 -15.55 -20.55 24.45
## [,21] [,22] [,23] [,24] [,25] [,26] [,27] [,28] [,29] [,30] [,31]
## COVER1 57.05 17.05 2.05 -7.95 -32.95 -32.95 -22.95 7.05 2.05 22.05 2.05
## COVER2 61.35 52.35 47.35 -22.65 -37.65 -37.65 -27.65 37.35 32.35 52.35 57.35
## COVER3 64.45 44.45 34.45 4.45 -35.55 -35.55 -15.55 -5.55 -15.55 54.45 44.45
## [,32] [,33] [,34] [,35] [,36] [,37] [,38] [,39] [,40] [,41]
## COVER1 -32.95 -32.95 -27.95 -12.95 -32.95 -32.95 -32.95 -17.95 62.05 7.05
## COVER2 -37.65 -37.65 -36.65 22.35 -37.65 -37.65 -37.65 17.35 -37.65 -2.65
## COVER3 -35.55 -35.55 -33.55 14.45 -35.55 -35.55 -35.55 14.45 4.45 -5.55
## [,42] [,43] [,44] [,45] [,46] [,47] [,48] [,49] [,50] [,51] [,52]
## COVER1 7.05 -17.95 -2.95 42.05 67.05 67.05 67.05 67.05 67.05 67.05 67.05
## COVER2 12.35 22.35 -7.65 47.35 62.35 52.35 57.35 57.35 61.35 -7.65 -32.65
## COVER3 4.45 -30.55 -20.55 39.45 64.45 49.45 64.45 64.45 64.45 59.45 59.45
## [,53] [,54] [,55] [,56] [,57] [,58] [,59] [,60]
## COVER1 -32.95 -27.95 47.05 47.05 47.05 7.05 -12.95 -31.95
## COVER2 -37.65 -32.65 52.35 57.35 52.35 17.35 2.35 -37.65
## COVER3 -35.55 -30.55 49.45 44.45 34.45 14.45 -30.55 -35.55
## attr(,"scaled:center")
## COVER1 COVER2 COVER3
## 32.95 37.65 35.55
Z_t_Z <- Z_t %*% Z
Z_t_Z
## COVER1 COVER2 COVER3
## COVER1 76734.85 60191.95 72989.65
## COVER2 60191.95 86335.65 70795.55
## COVER3 72989.65 70795.55 82812.85
N <- nrow(Z) # Number of observations (rows)
cov_matrix_manual <- (1 / (N - 1)) * Z_t_Z
# Display the result
print(cov_matrix_manual)
## COVER1 COVER2 COVER3
## COVER1 1300.591 1020.203 1237.113
## COVER2 1020.203 1463.316 1199.925
## COVER3 1237.113 1199.925 1403.608
p. 35
Covariance Decomposition Derivation
We are given the covariance expression for two variables, and :
Step-by-Step Derivation:
Start with the covariance formula:
We are looking at the covariance between two linear combinations of random variables:The general covariance formula for two linear combinations is:
In our case, the random variables are and .
Applying the formula:
Expanding the covariance expression:Since and are constants, we can take them outside of the covariance:
Simplifying the terms:
- (the variance of )
- (assuming and are independent)
- (assuming and are independent)
- (assuming and are uncorrelated)
Therefore, the expression simplifies to:
Conclusion:
The covariance between and is: